Tandem Ubiquitin Binding Entities (TUBEs)
Tandem Ubiquitin Binding Entities or TUBE technology is based on the ability to harness the strength of multiple UBDs (Ubiquitin Binding Domains), circumventing the need for immunoprecipitation of overexpressed epitope-tagged ubiquitin or the use of ubiquitin antibodies.
Need Help?

- How TUBEs Work
Antibody-based ubiquitin proteomics is expensive for large scale studies, and, moreover, ubiquitin antibodies sold by most suppliers are notoriously non-selective, leading to artifacts.
LifeSensors has developed technology to specifically isolate polyubiquitylated proteins from cell lysates and tissues. TUBEs are tandem ubiquitin binding domains (UBDs) that bind polyubiquitin with Kds in the nanomolar range.
High Affinity Binding
Kds in the nanomolar range for superior specificity
Protein Protection
Prevents deubiquitylation and degradation
No Inhibitors Required
Works effectively without additional blocking agents
Key Mechanism
TUBEs have been demonstrated to protect ubiquitylated proteins from both deubiquitylation and proteasome-mediated degradation, even in the absence of inhibitors normally required to block such activities. This revolutionary approach eliminates common artifacts and provides more accurate results in ubiquitin research.
Industry-Leading Performance: Lysine Specfic Targeting
Our Tandem Ubiquitin Binding Entities have transformed the ubiquitin community due to the powerful ability to selectively capture polyubiquitylated proteins. The combination of superior affinity, protective properties, and cost-effectiveness makes TUBEs the gold standard in ubiquitin research.
- Advantages of Choosing LifeSensors' TUBEs
Significantly more affordable compared to alternative technologies while maintaining superior performance and reliability for large-scale studies.
Exceptional binding affinity in the high nanomolar range ensures precise and reliable capture of polyubiquitylated proteins with minimal background.
Unique protective properties prevent unwanted deubiquitylation and proteasomal degradation, preserving sample integrity throughout your workflow.
Versatile platform spanning beyond the TPD industry, enabling diverse research applications from basic science to cancer research to drug discovery.
High-Throughput Screening Platform
Utilizing pan-selective and chain-selective TUBEs with nanomolar affinity, LifeSensors has developed a variety of assays and services utilizing TUBE technology. Currently, TUBEs are being used by LifeSensors to develop an HTS that elevates PROTAC efficiency aiding TPD (Targeted Protein Degradation) based therapeutics.
TUBE Pulldowns
TUBEs, such as UM501M are commonly used for isolation and pulling down of ubiquitinated proteins, allowing for a range of applications such as Mass Spec Proteomics, Imaging, or more traditional western blot assays. The detailed experimental procedure references a magnetic TUBE, UM501M, for the purposes of pulling down ubiquitinated proteins.
Isolation of Ubiquitinated Proteins
Enables reliable capture of ubiquitinated proteins from complex samples.
Mass Spec Proteomics Compatible
Supports downstream analysis for proteomics workflows.
Detailed Experimental Procedure
LifeSensors has support for a step-by-step manual for accurate pulldown execution.
- Resources
LifeSensors TUBEs: Tandem Ubiquitin Binding Entities
Technology Overview and Presentation
PROTAC® Drug Discovery with TUBEs
Download our Published Paper
White Paper and Technology
Applications
UM501M Magnetic TUBE Manual
Magnetic TUBE Pulldown Procedure
- E3 Ligase Assays
Quickly distinguishing true hits from false positives, TUBE-based assays develop structure-activity relationships, and establishing rank order potency from purified enzymes to cellular models are key steps to success.
LifeSensors brings together the tools and expertise necessary to overcome the many pitfalls in the E3 ligase field.
Hit Validation
Rapid identification and confirmation of true positive compounds
SAR Development
Comprehensive structure-activity relationship analysis
Potency Ranking
Accurate rank order determination across model systems
- PROTAC® Discovery Platform using TUBEs
High-throughput tools for monitoring both polyubiquitylation and degradation of a target protein. Measuring ubiquitylation of a protein in a plate-based format accelerates PROTAC-based drug discovery. TUBEs form the cornerstone of this platform, where unmodified proteins can engage in a natural environment relative to luciferase type assays which can be prone to artifacts.
LifeSensors has developed many high-throughput 96-well and 384-well plate-based assays in order to address the needs of the PROTAC and the TPD field for reliable, sensitive assays to study the impact of a molecule on ubiquitination or a target. These assays are great for hit confirmation and can also be used for Mass Spec Proteomics.
Furthermore, we have several platforms that enable rapid, quantitative monitoring of in vitro as well as cellular ubiquitylation allowing for an accurate picture of how a target is being degraded, valuable for toxicology profiles.
Quantitative Monitoring
Real-time tracking of ubiquitylation levels
Plate-Based Format
High-throughput screening capability
In Vitro & Cellular
Comprehensive validation across systems
Molecular Glue Discovery Platform using TUBEs
PROTACs have opened new vistas of targeted protein degradation. However, complex medicinal chemistry, warhead optimization, linkerology, and inefficient methods to analyze degradation of the target proteins in cells has hampered its progress. Historical data has shown that optimized molecular glues (MGs) perform the same function as PROTACs.
Molecular glues have a small molecular weight, and are discovered by traditional screens. Potent and selective glues can be designed with simple chemistry. LifeSensors provides a plethora of tools to discover molecular glues based on our TUBE platform as well as supporting reagents. Measuring ubiquitylation of a protein in a plate-based format accelerates MG-based drug discovery.
We have developed several platforms that enable rapid, quantitative monitoring of in vitro as well as cellular ubiquitylation.
Key Advantages
Plate-Based Format
High-throughput screening capability
Small Molecular Weight
Enhanced cell permeability and bioavailability
High Selectivity
Potent and specific target engagement
- 1-10 nM Affinity
- All Chain Types
- Biomarker Discovery
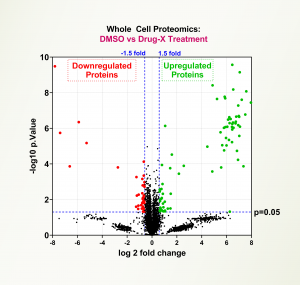
Mass Spec Proteomics
LifeSensors’ TUBEs bind to all polyubiquitin chains with 1-10 nM affinity, overcoming major problems common to mass spectrometry proteomics.
The combination of the TUBE-based affinity technology and targeted mass spectrometry is the most powerful way to detect the alterations in PTMs and identify signatures for research and biomarkers.
Platform Capabilities
PTM Detection
Sensitive identification of post-translational modifications
Signature Identification
Discovery of disease-specific ubiquitylation patterns
Biomarker Discovery
Identification of therapeutic and diagnostic targets
Imaging Solutions
TAMRA-TUBE 2 has a single TAMRA fluorophore (Exc. = 540 nm, Emm. = 578 nm) attached to the fusion tag of TUBE 2 (Cat. No. UM202).
Since the fluorophore is attached to the tag, it does not affect binding of the tandem ubiquitin binding domains to polyubiquitin chains allowing for utilization of imaging techniques to study natural ubiquitination.
Other fluorophores and conjugations are available upon request.
- Compatible Techniques
- Confocal Microscopy
- Widefield Fluorescence
- Flow Cytometry
- High-Content Screening
Platform Capabilities
PTM Detection
Sensitive identification of post-translational modifications
Signature Identification
Discovery of disease-specific ubiquitylation patterns
Biomarker Discovery
Identification of therapeutic and diagnostic targets
No Binding Interference
Tag-attached fluorophore preserves TUBE functionality
Live Cell Imaging
Real-time visualization of ubiquitination dynamics
- Related Products
- Pan-Selective TUBEs
Product / Tools Name
SKU
Categories
Add to cart
- K48 TUBEs
Product / Tools Name
SKU
Categories
Add to cart
- K63 TUBEs

