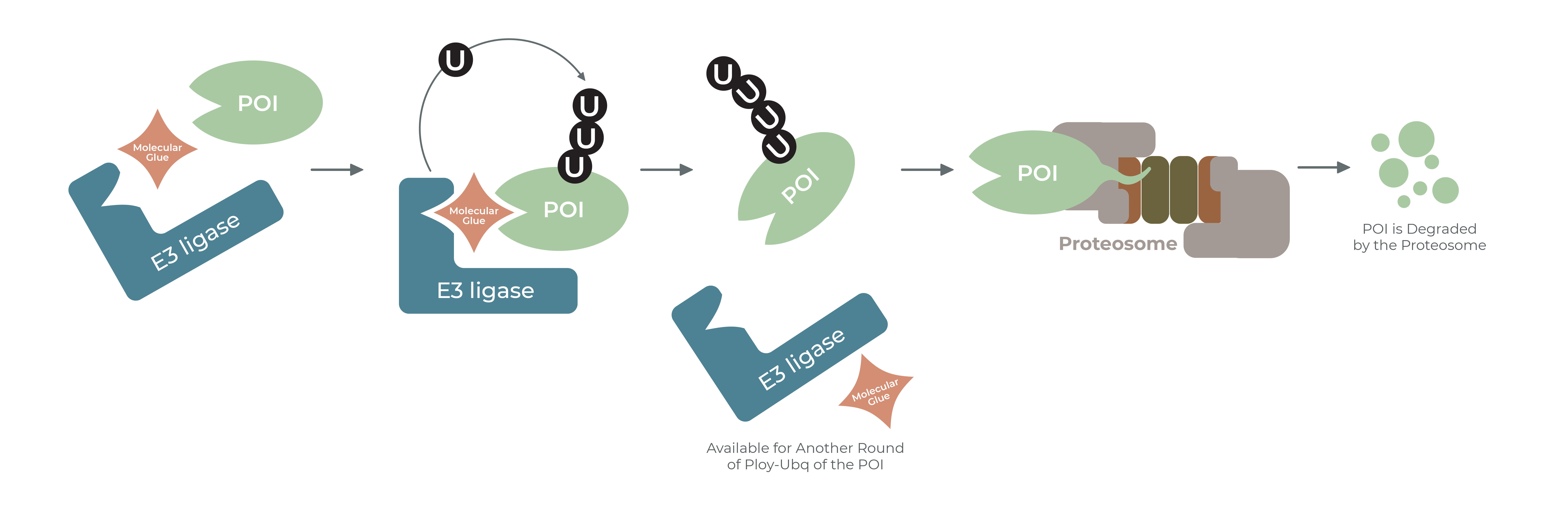
PROTACs have opened new vistas of Targeted Protein Degradation. However, complex medicinal chemistry, warhead optimization, linkerlogy, and inefficient methods to analyze degradation of the target proteins in cells has hampered its progress. Historical data has shown that optimized molecular glues (MGs) perform the same function as PROTACs. Molecular glues have a small molecular weight, discovered by traditional screens. Potent and selective glues can be designed with simple chemistry. LifeSensors provides a plethora of tools to discover molecular glues.
There are mainly two types of molecular glues. The first class of glues are those that bind to an E3 ligase to change its conformation so that it binds and ubiquitinates a neo-substrate. The second class of glues will bind to a target protein, change its conformation such that the structure is altered, exposing lysines to be ubiquitinated by neighboring ligases. The net effect is the same – the target protein is ubiquitinated and degraded. Depending upon the type of ligase employed for ubiquitination, if lysine 48 chains are built on the target protein, the proteasome will recognize the target and degrade it. If lysine 63 chains are built on the target protein, the protein may be trafficked to another compartment with a loss of function. However, lysine 63 chains most likely promote autophagy, and the protein will be degraded by lysosomal degradation mechanisms. LifeSensors has a variety of tools to discover and tease out the mechanism of molecular glue mediated ubiquitination and degradation.