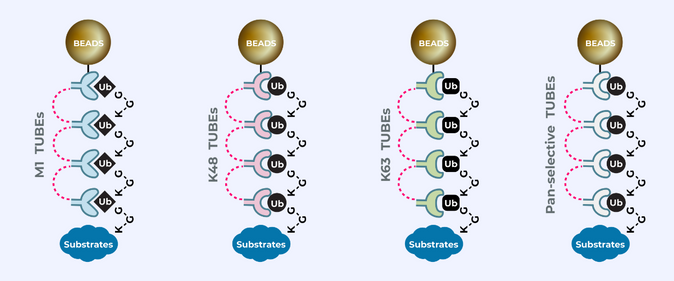
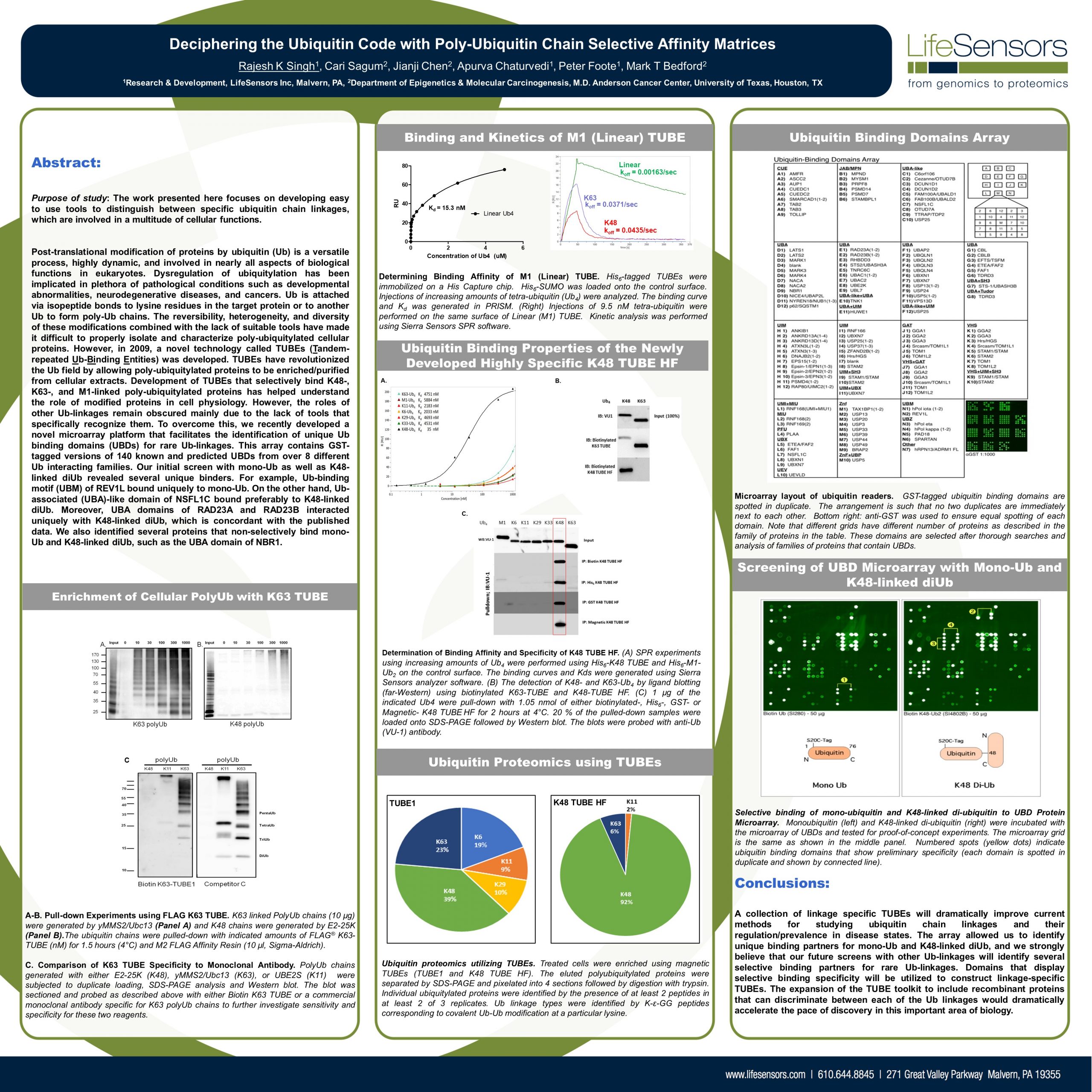
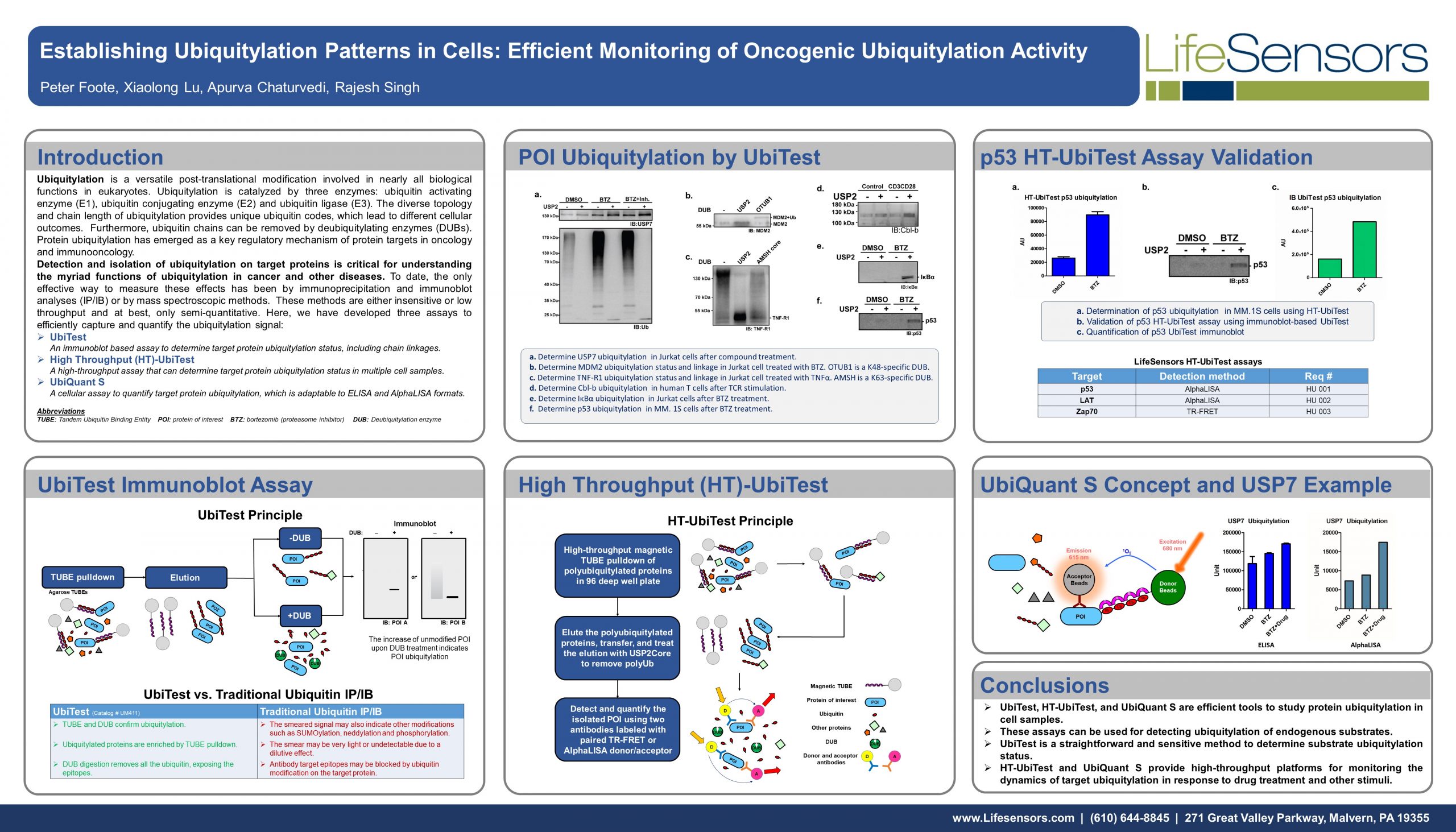
UbiTest: Detection of polyubiquitinated proteins using TUBE based screening platforms by Gauthami S. Jalagadugula The ubiquitin proteasome system (UPS) is a highly regulated mechanism that controls many cellular processes through…

UbiTest: Detection of polyubiquitinated proteins using TUBE based screening platforms by Gauthami S. Jalagadugula The ubiquitin proteasome system (UPS) is a highly regulated mechanism that controls many cellular processes through…

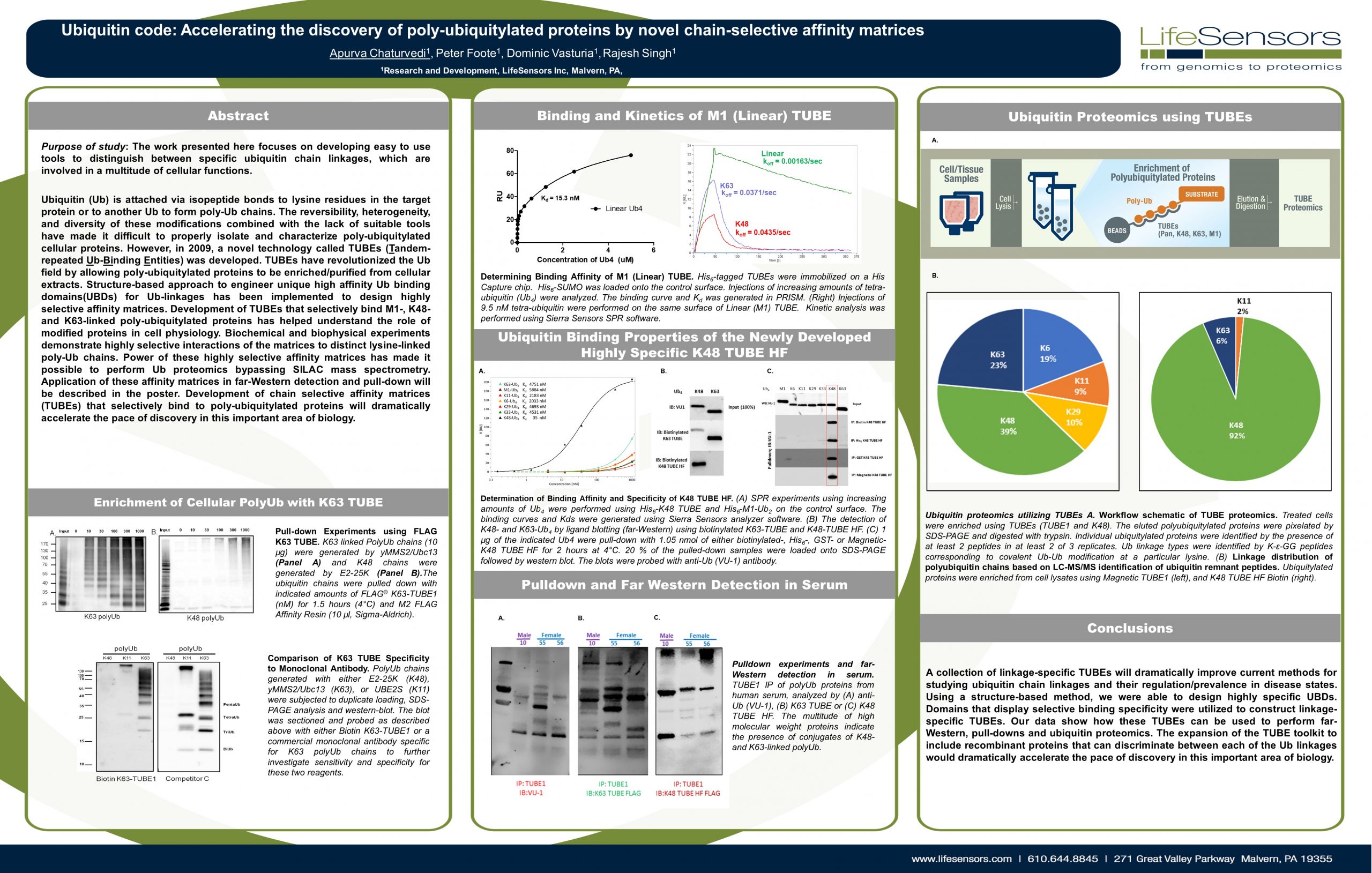
Hijacking the natural mechanism of the Ubiquitin Proteasome System (UPS) is emerging as a powerful medium in drug discovery through which Dr. Kadimisetty uses “TUBEs” as ubiquitin-binding entities to monitor…

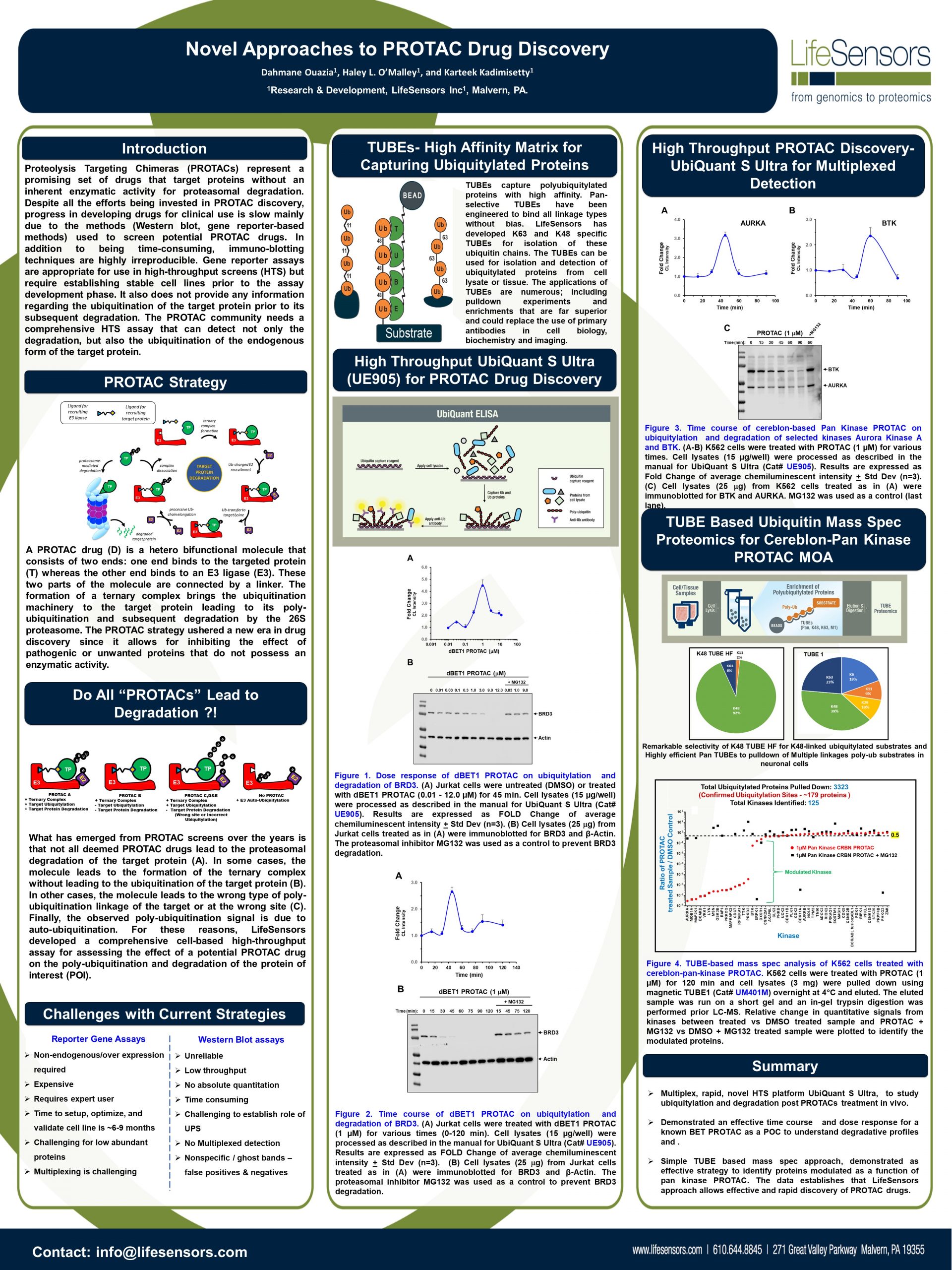
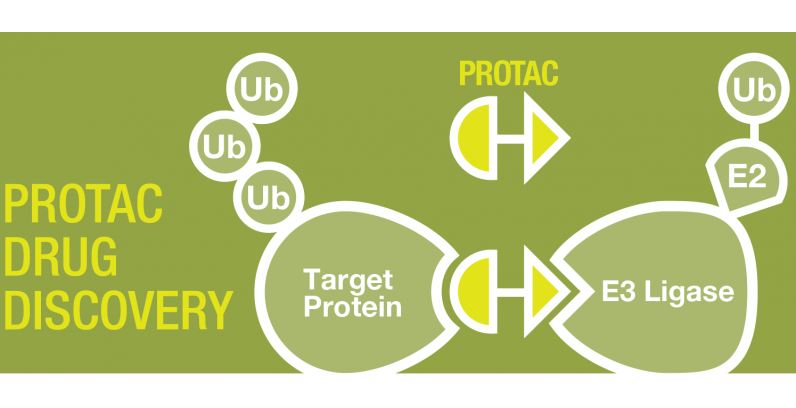
Discovering Unique Ubiquitin E3 Ligase Ligands to Unlock Targeted Protein degradation Gauthami Jalagadugula, Alexandra Russell1+, Kenneth Wallace1+, Janelle Pedroza and Abdul Haseeb1 * Introduction PROteolytic TArgeting Chimeras (PROTACs) have emerged…

PROTACs and Molecular Glues: High-Throughput Quantitative Assays for Rationale Design Authors: Janelle Pedroza1, Kenneth Wallace1, Derrick Padykula, Alexandra Russell1, and Karteek Kadimisetty*1 Introduction PROTACs have emerged as new class of…


271 Great Valley Parkway
Malvern, PA 19355
Phone: 610.644.8845
Fax: 610.644.8616