Tandem Ubiquitin Binding Entities (TUBEs) are industry-leading tools to detect and enrich/purify poly-ubiquitylated proteins from cell lines, tissues, and organs. Pairing this sample processing step with LC/MS analysis, generates a reproducible survey of the ubiquitome. We have worked with clients to investigate ubiquitin levels as a response to drug dosing, genetic knockouts, disease states, and others.
2. What are the disadvantages of traditional methods?
Traditional Mass spec proteomics methods do not work well for the ubiquitin proteome system. Ubiquitylation of proteins is very dynamic and sample preparation to preserve pattern of ubiquitylation is critical. Above all methods used to enrich ubiquitylated peptides by di-Gly-Lys antibody fail to capture all the ubiquitylated proteins. Because of this, traditional approaches have proven to be irreproducible.
3. What can I expect to get out of a mass spec service?
Clients receive their processed data in an excel file, as well as a summary report. We include graphical representations of ubiquitylation tends of your proteins. You will receive professional support when analyzing your data. See example report.
- Written report with summary of the TUBE based mass spec proteomics data, with sample prep protocols and LC-MS methods implemented for given samples along with sample description.
- Total Ubiquitinated identified will be presented in a list, as well as being organized on a distribution chart to show proportions.
- Total peptides identified will be presented in a list with length and mass information.
- Additional bioinformatic analysis for getting functional perspective of Proteomics data like DE analysis, cluster analysis and pathway & network analysis are available upon request at addition costs.
4. What is the typical work flow?
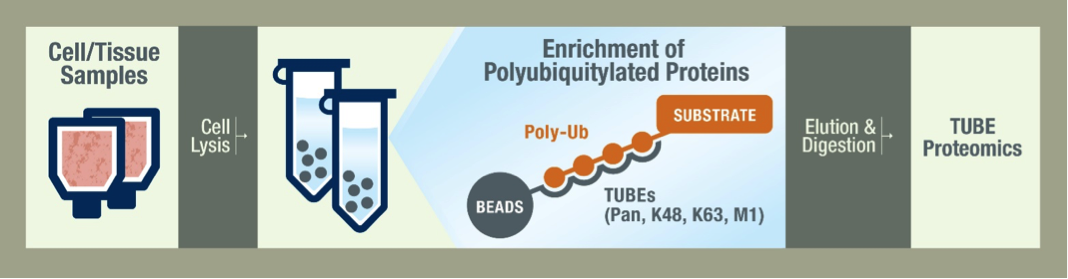
Your job ends at the shipment of frozen cells or tissue. Our scientists lyse your cells with a cocktail of inhibitors to protect the ubiquitylated species. The samples are then prepared using the TUBEs to pulldown the ubiquitylated proteins. Pan selective TUBEs are recommended for a global view of the ubiquitiome, but we also offer K48 and K63 linkage-specific TUBEs for specialized studies. Proteins are then separated on a gel by SDS-PAGE and are stained for analysis. After data is collected by LC-MS/MS we begin data analysis by running bioinformatic searches. The depth of our analysis depends on your needs and budget and we are happy to work with you to decide the best methods.
5. What is the required amount of starting material?
We require sample of 1mg-5mg of total protein for this analysis. Depending on your cell type this will equate to pellets of approximately 50 million cells. If you are using tissue samples, we can discuss sample sizes that would be sufficient.
6. Do you recommend duplicates? Triplicates? ?
We recommend submitting at least two samples for each condition for a more robust analysis. If you are looking to publish this data, duplicates are typically enough for this type of experiment. Ensure that you design your experiment with the proper controls, and in duplicate in order to have enough data to draw meaningful conclusions.
7. What is the turnaround time?
We are typically able to deliver a report after 2-3 weeks upon receipt of the samples. Depending on the number of projects at any given time this is subject to change. Please contact us to get a more accurate turnaround time for your specific project.
8. What type of control samples do you recommend?
The controls for each experiment depend on the question being asked, but there are some guidelines that you might find useful. If you are looking into the effects of a drug or other compound, we recommend submitting a negative control. For genetic studies the negative control may be a wildtype sample. The use of inhibitors is another important control in this type of study. We recommend submitting samples treated with proteasome inhibitors, such as MG-132 to accumulate ubiquitylated proteins. We are happy to help you design a robust experiment around your specific research question, please contact us.
9. Is this analysis quantitative?
Yes, the TUBE-based mass spec analysis is quantitative. The intensities reported describe the abundance of that peptide or protein. These values account for a loading control, as well as the size of the proteins.
