TUBE-Based Mass Spectrometry Proteomics
Advanced enrichment and analysis of polyubiquitylated proteins using our proprietary TUBE technology combined with state-of-the-art mass spectrometry
Need Help?

- TUBE-Based Mass Spectrometry Proteomics
Tandem Ubiquitin Binding Entities (TUBEs) are powerful reagents for enrichment of polyubiquitylated proteins. At LifeSensors, we have used this remarkable technology of TUBEs with our mass spectrometry expertise to provide our customer with a quick and easy way to perform both qualitative and quantitative proteomics.
Upon completion of the analysis, you will receive a list of peptides and corresponding proteins that have been identified. We will categorize the ubiquitylated proteins and display the results on several plots to make your analysis easier. This information will help you publish your results or plan your next assay.
Quick Turnaround
Comprehensive Analysis
Expert Support
- Platform Resources
LifeSensors DUBs Screening & Profiling Platform
Learn more about our comprehensive DUBs screening and profiling platform
- Workflow Schematic
- Project Design
Experimental Design Consultation
We are here to help you from the beginning, and are happy to sit down and help you design the best experiment to address your specific questions.
Competitive Pricing
We have competitively priced our analysis and offer custom services with the level of analysis that matches your needs and budget.
Fast & Professional Reports
We strive to deliver you accurate and professional reports quickly. From submission of samples, reports are typically delivered in 2-4 weeks depending on the complexity of project.
- Proprietary TUBE Technology
TUBE-based enrichment of polyubiquitylated proteins has proven to be key for the progress of ubiquitin proteomics. LifeSensors has developed the proprietary technology to identify cellular proteins that are ubiquitylated using TUBE-based proteomics.

TUBE Enrichment Process
- Lyse cells
- Wash non-specific proteins
- Elute proteins
- Protease digestion
Resources Available
Download Sample Report PDF
- Chain Selectivity
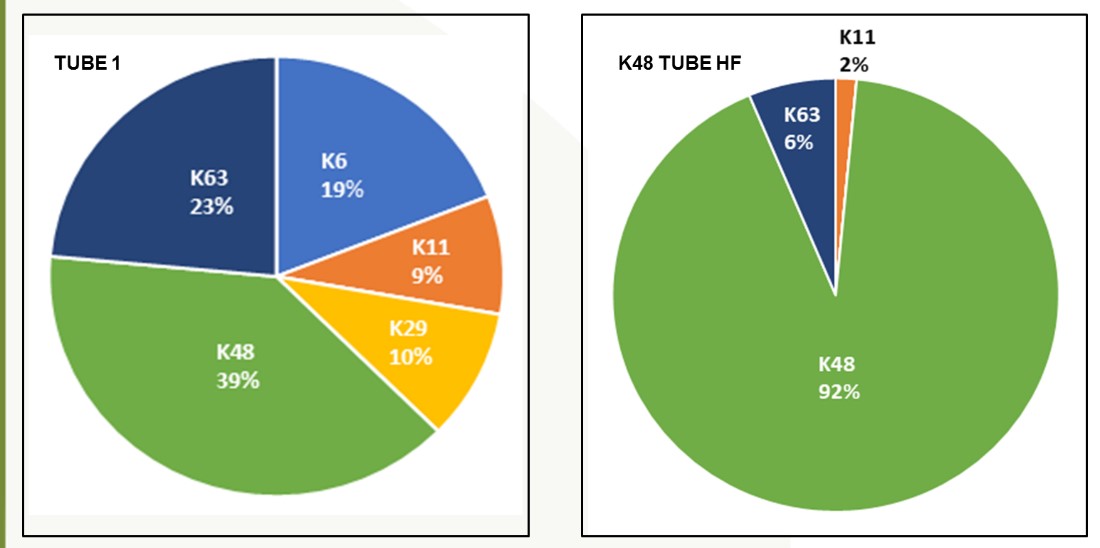
- Experimental Data
In-house Data
Below are examples of data generated using TUBE-based mass spec technology with pan-selective TUBEs. Here, the ubiquitinated proteins have been differentiated from whole cell lysate. This methodology is sensitive enough to detect the number of ubiquitin sites on a given protein.
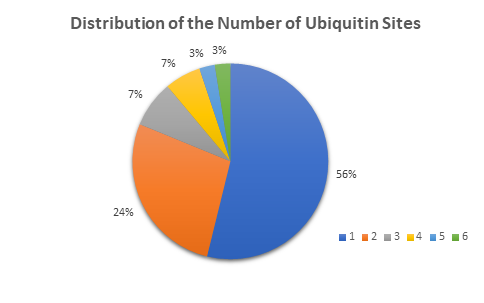
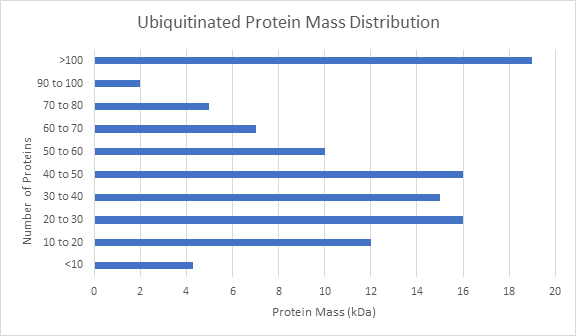
When studying the effects of a drug or genetic knockout, global shifts in the ubiquitome is a good place to start your analysis. Other global analysis provided by LifeSensors includes protein mass distribution of all proteins as well as ubiquitinated proteins.
- Published Data
- Silva et al. Study
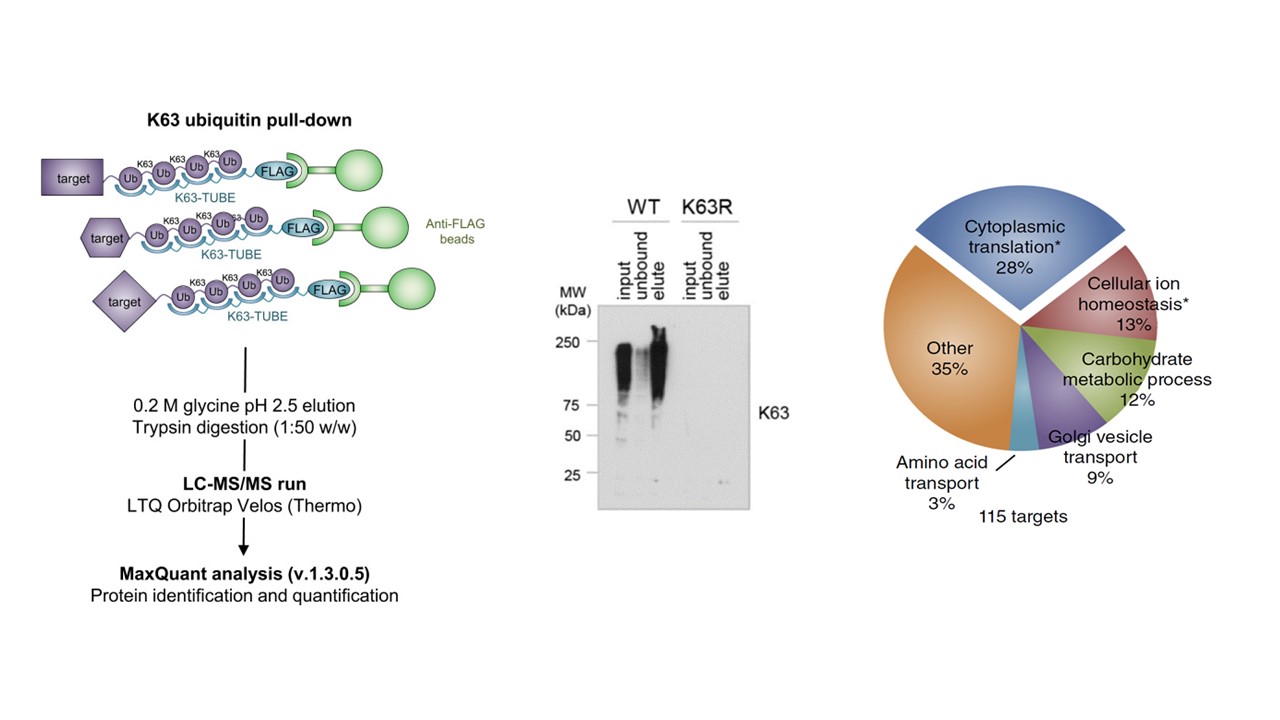
- Mata-Cantero et al. 2016
In 2016, Mata-Cantero et al. used TUBE-based mass spectrometry to identify major components of the ubiquitin proteome of both Plasmodium falciparum and its host during different life stages.
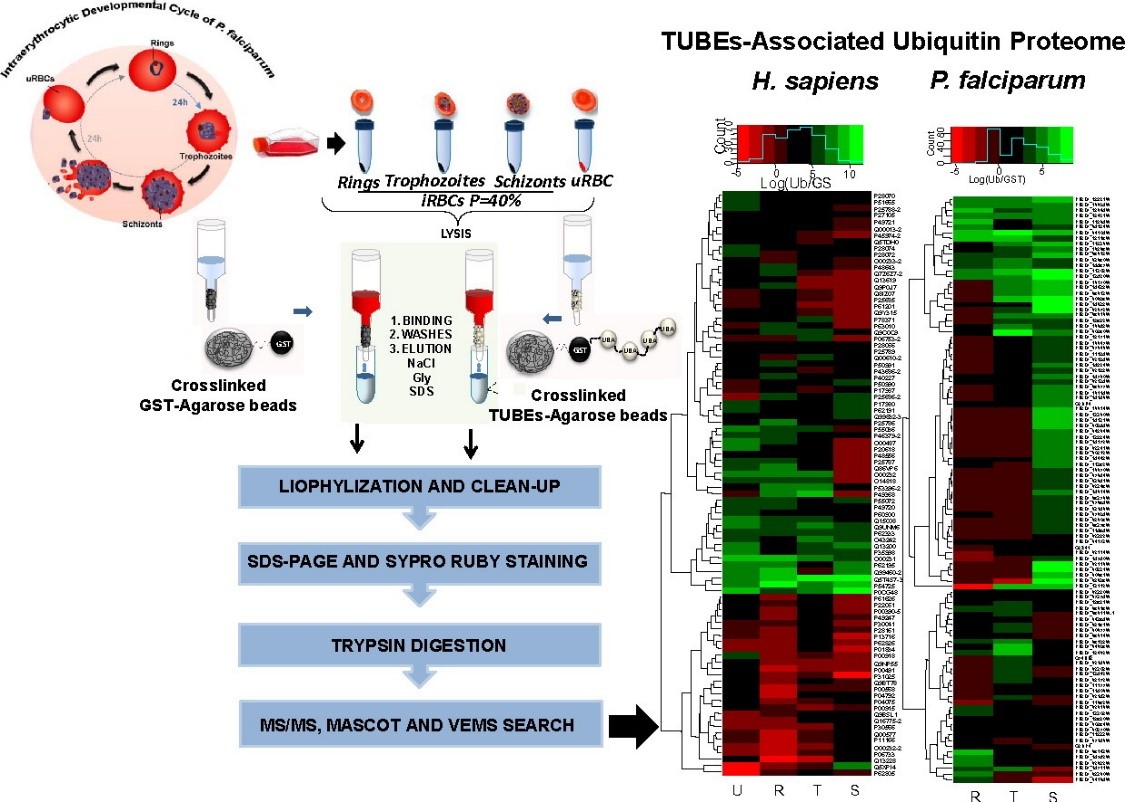
Intraerythrocytic Developmental Cycle of P. falciparum is shown. Synchronized P. falciparum iRBC at 40% parasitaemia from rings, trophozoites and schizonts stages were collected and frozen. TUBE enriched proteins from iRBC at different stages and uRBC were captured using TUBEs or GST (control) previously crosslinked with DMP to agarose beads. After exhaustive washes, proteins captured were eluted, cleaned by precipitation and resolved by electrophoresis (PAGE). Bands with proteins were analyzed by LC-MS/MS.
