E3 Ligase Drug Discovery, Profiling & Screening
Advanced reagents and assays for comprehensive E3 ligase screening and profiling solutions
Need Help?

- Overview
Quickly distinguishing true hits from false positives, developing structure-activity relationships, and establishing rank order potency from purified enzymes to cellular models are key steps to success. LifeSensors brings together the tools and expertise necessary to overcome the many pitfalls in the E3 ligase field.
LifeSensors’ small molecule library contains a collection of ligase-centric compounds, which can be employed as a gold standard for your ligase drug discovery efforts. Further, LifeSensors has the tools to quickly exclude off-target hits through a variety of complementary biochemical and biophysical validation assays. In E3 ligase drug discovery, connecting the dots is particularly important.
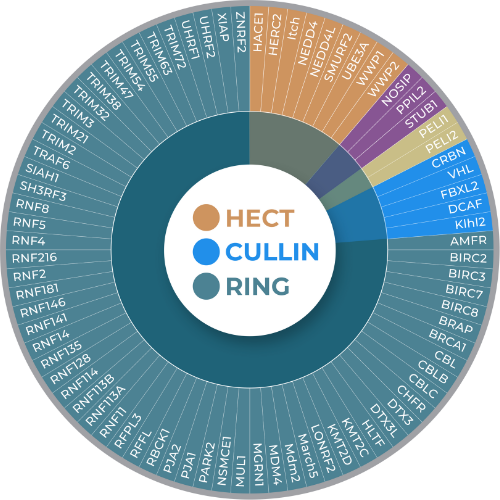
Understanding E3 Ligase Classifications
Important introduction to E3 Ligases and new classifications beyond the standard familiy format (2025)
E3 Ligase Key Services for Discovery
HTS Biochemical Assays
LifeSensors offers a range of powerful assays that can be run in-house, from gel and TR-FRET assays, to auto-ubiquitination and substrate ubiquitination assays
Substrate & E3 Finder and Profiler
Taking advantage of our HTS assays discover how a ligase of interest binds and ubiquitinates a target.
Cell-Based Assays
With multiple in-house developed ELISA-based assays, LifeSensors is uniquely suited to evaluate post-translation modifications in intact cells
In Vitro Biophysical Assays
Complementary biophysical tools widely used in E3 ligase drug discovery to characterize small-molecule binders
- Key Methodologies
E3s are Rarely Discovered With a Single Assay
We are here to help you from the beginning, and are happy to sit down and help you design the best experiment to address your specific questions, as well as answer questions that matter for your project's completion
Industry Tested
As a CRO that's been in the TPD space since the beginning, we're familiar with the long developmental time and have tested projects that are currently in the clinic. We can tell you the assay selection you would likely need in order to discover, test, and validate your targets
Competitive Pricing
We have competitively priced our analysis and offer custom services with the level of analysis that matches your needs and budget. We're flexible but also want to make sure the assay can provide the data it needs to advance your project
Fast & Professional Reports
We strive to deliver you accurate and professional reports quickly. From submission of samples, reports are typically delivered in 2-4 weeks depending on the complexity of the project
E3 ligases remain among the most difficult enzyme targets to assay and discover potent therapeutic applications. This is possibly due to their dependence on upstream E1 and E2 enzymes for the activation and transfer of ubiquitin. As a result, traditional assays for E3 ligases are complex and prone to off-target false positives. At LifeSensors, we have developed reagents and assays encompassing both traditional and non-traditional approaches for E3 ligase screening and profiling. Quickly distinguishing true hits from false positives, developing structure-activity relationships, and establishing rank order potency from purified enzymes to cellular models, are key steps to success. LifeSensors brings together the tools and expertise necessary to overcome the many pitfalls in the E3 ligase field.
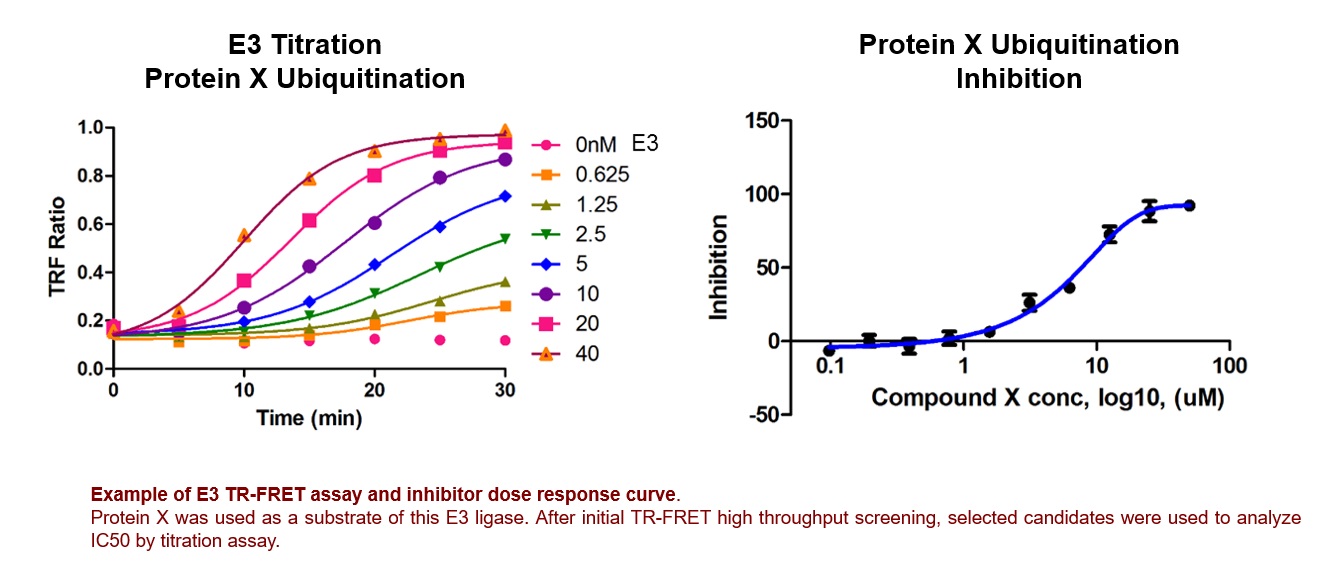
Our E3 TR-FRET detection system utilizes our TUBE technology to monitor E3 ligase activity by fluorescence signal. This assay involves donor-labeled TUBEs that bind to acceptor-labeled polyubiquitin chains synthesized by the target E3 ligase. When donor-labeled TUBEs come in close proximity to polyubiquitin chains containing acceptor-labeled ubiquitin will yield in a FRET signal. This signal can be monitored over time in a homogenous, high-throughput format, making it ideal for small molecule screening to find tool compounds for E3 ligases.
Service Highlights
- High-throughput screening using compound libraries to discover novel inhibitors/activators/binders for E3 ligases
- Automated liquid handling amenable to large library screening
- Access to 50+ E3 ligases for performing discovery and selectivity studies in rapid homogenous assay format
- Access to customized collection of ligase centric small molecule libraries for using as reference in ligase drug discovery efforts
- Ultrasensitive with robust signal-to-noise powered by TUBE technology
- Extensive experience in UPS related drug discovery – complementary biochemical and biophysical validation studies to exclude off-target hits
Besides aiding drug discovery, E3 ligase screening and profiling services at LifeSensors also offer identification of E2 pairs for the E3 ligase of interest, identification of the nature of polyubiquitination on the E3 ligase and protein of interest, in order to gain insights into signaling mechanisms using chain selective TUBE technology.
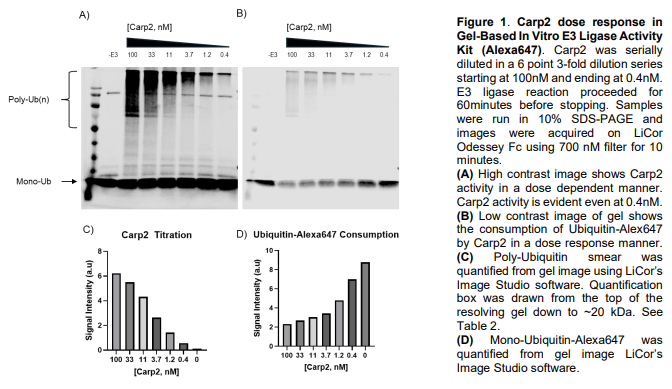
Gel-based assays for evaluating E3 ligases rely on in vitro ubiquitination reactions reconstituted with E1, E2, a chosen E3 ligase, ubiquitin, and ATP, followed by SDS-PAGE and Western blotting to visualize ubiquitination. These assays are commonly used to assess E3 activity, auto-ubiquitination, substrate modification, E2 specificity, and the effects of mutations or inhibitors. Readouts appear as molecular-weight shifts or smears corresponding to mono- or polyubiquitinated species, detected with antibodies against ubiquitin, the E3, or the substrate. While gel-based approaches are low-cost and mechanistically informative, they are semi-quantitative and limited in throughput and chain-type resolution, often serving as a foundation for more advanced biochemical or mass spectrometry-based analyses.
LifeSensors offers a range of technologies and assays for gel-based evaluation, off the shelf, but also provides customer focused development and design in order to assist with project testing.
Service Highlights
- TUBE-based technology allows for a clear picture of total or lysine specific ubiquitination
- Quantitative measurement utilizing UbiQuant, UbiTest, and other well validated ubiquitin-based assays
- Assays available to be taken in-house to advance the project on additional targets
- Gel-based readouts improve turnaround time relative to western blot measurements
- Autoubiquitination, substrate ubiquitination, and activity assays can deconvolute hits from E3 screens
- Extensive experience in UPS related drug discovery – complementary biochemical and biophysical validation studies to exclude off-target hits
Besides aiding drug discovery, E3 ligase screening and profiling services at LifeSensors also offers identification of E2 pairs for E3 ligase of interest, identification of the nature of polyubiquitination on E3 ligase and protein of interest in order to gain insights into signaling mechanisms using chain selective TUBE technology.
In Vitro Ubiquitination
Recreate the ubiquitination cascade to evaluate polyubiquitination with TUBEs. Determine activity and chain length
Substrate Ubiquitination
Take advantage of LifeSensors protein expression to work with purified substrates and proteins for biologically meaningful data
Custom Assay Design
With many years combined experience working within the Ubiquitin Proteasome System, LifeSensors has the capability to custom design and provide procedural knowledge to take those assays internally
Auto-Ubiquitination
RING and HECT ligases frequently ubiquitinate themselves for a fast readout of E3 catalytic capability. Fast and a great place to evaluating an E3s
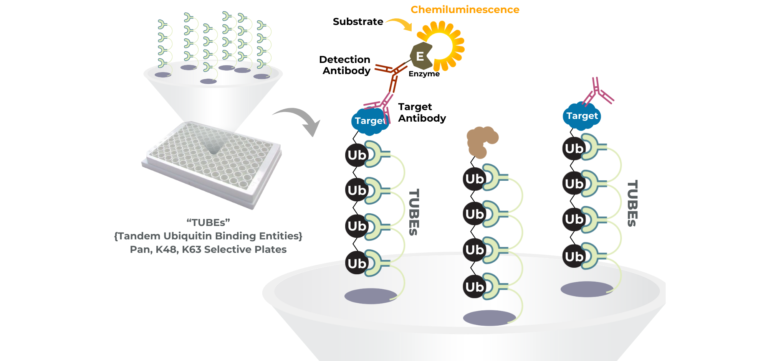
Unlike binding or proximity assays, LifeSensors’ high-throughput screening platforms measure what truly matters for PROTACs and molecular glues: target protein ubiquitination. Powered by proprietary Tandem Ubiquitin Binding Entity (TUBE) technology, these assays capture and quantify polyubiquitinated proteins with exceptional sensitivity and specificity in scalable microplate formats.
TUBE-based HTS assays enable rapid screening and ranking of PROTACs and molecular glues based on their ability to drive ubiquitination through native E3 ligase pathways. Pan- and linkage-selective TUBEs provide additional mechanistic insight by distinguishing degradation-relevant ubiquitin chains (such as K48) from non-degradative signaling events.
By translating ubiquitination biology into a robust, plate-based assay format, LifeSensors’ TUBE-powered HTS platforms help teams move beyond surrogate endpoints and confidently advance the most effective targeted protein degraders.
Service Highlights
- Well validated assays performed in-house to create a detailed workflow for target exploration
- Assays can be taken in-house to advance the project on additional targets or screen expansion
- Measuring natural ubiquitination via TUBEs avoids artifacts associated with more commonly used tag based assays
- Mechanistic profiling of ubiquitin chain topology
- SAR-driven ranking of PROTAC and molecular glue candidates
Chemiluminescent Ubiquitination Assays
Explore the quantitative interaction of a known E3 ligase with unknown substrates, or screen for E3s that target a substrate to determine E3 substrate pairs
TUBE Mass Spec Proteomics
The most detailed evaluation of a target's ubiquitination status coupled with lysine specific TUBEs as a key preparation reagent allowing a high level of detail, important for evaluating lysine specific ubiquitination
Our substrate and E3 finder platform provide a powerful tool for molecular glue discovery by directly measuring E3 ligase activity and substrate ubiquitination in a controlled, quantitative setting. Molecular glues function by stabilizing or inducing interactions between an E3 ligase and a target protein, so detecting enhanced ubiquitination of a candidate substrate is a key readout of glue activity. This platform leverages proprietary polyubiquitin capture reagents to sensitively detect ubiquitination events, allowing researchers to determine whether a small molecule promotes E3-dependent ubiquitin transfer to a target of interest.
The chemiluminescent 96-well plate format enables parallel evaluation of multiple substrates, E3 variants, and compound conditions, making it suitable for screening libraries of candidate molecular glues and quantifying their relative potency. When a high-quality substrate antibody is available, the assay provides a specific, direct readout; when antibodies are lacking, mass spectrometry–based proteomics (via the E3 ID service) can identify novel ubiquitination events induced by candidate compounds. Together, these capabilities allow researchers to identify potential molecular glues, confirm their mechanism of action, and prioritize leads for further biochemical or cellular validation.
Service Highlights
- A deep exploration of the substrates engaged with a target E3 ligase to identify targets
- Well validated assays performed in-house to create a detailed workflow for target exploration
- Assays can be taken in-house to advance the project on additional targets or screen expansion
- Colorimetric readouts improve turnaround time and better quantification relative to western blot measurements
- Substrate ubiquitination, E3 ID, and activity assays, can deconvolute hits from E3 screens
- Flexible investigation based on current research, methodologies, and required level of detail
Chemiluminescent Ubiquitination Assays
Explore the quantitative interaction of a known E3 ligase with unknown substrates, or screen for E3s that target a substrate to determine E3-substrate pairs
TUBE Mass Spec Proteomics
The most detailed evaluation of a target's ubiquitination status coupled with lysine specific TUBEs as a key preparation reagent allowing a high level of detail, important for evaluating lysine specific ubiquitination
LifeSensors offers a multitude of cell-based services in order to convert biochemical findings into a cellular context. TUBEs allow for capture and quantification of ubiquitin in live cells, bacteria, yeast, mammalian, and plant cells, opening the door for drug discovery, target validation, and mechanistic studies without relying entirely on immunoprecipitation and western blots.
These assays are very well validated for over a decade and offer important information regarding ubiquitination, deubiquitinase performance, substrate performance, E3 activity all in response to the introduction of a treatment molecule, PROTAC, molecular glue, or an induced protein stabilizer.
Cellular Assays: an Overview
- PROTAC and molecular glue ubiquitination assays
- Quantitative measurement utilizing UbiQuant, UbiTest, and other well validated ubiquitin-based assays
- Assays available to be taken in-house to advance the project on additional targets
- Gel-based readouts improve turnaround time relative to western blot measurements
- Autoubiquitination, substrate ubiquitination, and activity assays can deconvolute hits from E3 screens
- Extensive experience in UPS related drug discovery – complementary biochemical and biophysical validation studies to exclude off-target hits
Besides aiding drug discovery, E3 ligase screening and profiling services at LifeSensors also offer identification of E2 pairs for the E3 ligase of interest, identification of the nature of polyubiquitination on the E3 ligase and protein of interest, in order to gain insights into signaling mechanisms using chain selective TUBE technology.
In Vitro Ubiquitination
Recreate the ubiquitination cascade to evaluate polyubiquitination with TUBEs. Determine activity and chain length
Substrate Ubiquitination
Take advantage of LifeSensors protein expression to work with purified substrates and proteins for biologically meaningful data
Custom Assay Design
With many years combined experience working within the Ubiquitin Proteasome System, LifeSensors has the capability to custom design and provide procedural knowledge to take those assays internally
Auto-Ubiquitination
RING and HECT ligases frequently ubiquitinate themselves for a fast readout of E3 catalytic capability. Fast and great place to start evaluating E3s
- Thermal Shift Assays
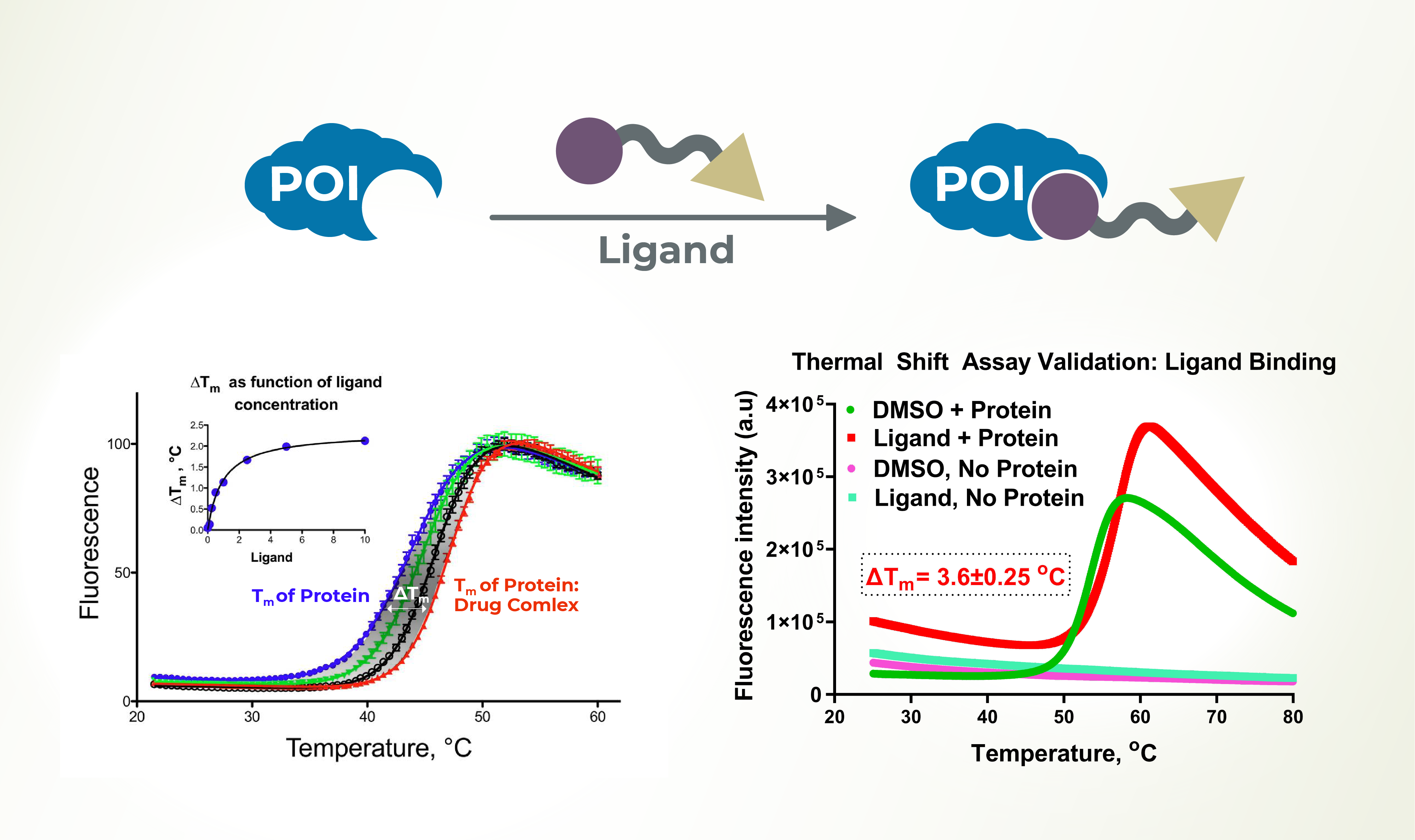
Generating experimental data for ligands binding to target proteins of interest, as well as determination of ligand binding affinity (Kd), is essential for drug discovery. The thermal shift assay is a quick and easy method to generate ligand-protein binding data and is widely employed across several target proteins and compounds, including natural products. LifeSensors has established a robust protein thermal shift assay platform and offers services to facilitate ligand discovery and characterization for clients. Together with our proprietary protein expression and purification technology, LifeSensors can help you discover ligands for difficult to express target proteins.
Service Highlights
- Label-free technology is amenable to an HTS
- Independent of catalytic activity
- Characterization of small molecule and PROTACs binding to target proteins
- Facilitates identification of E3 and DUB ligands
- Surface Plasmon Resonance
Generating experimental data for ligands binding to a target protein of interest as well as determination of the ligands binding affinity (Kd), is essential for drug discovery. SPR is a label-free optical technique that measures interactions in real time. A target of interest (ligand) is immobilized on a thin gold-coated glass chip while a second molecule (analyte) is flowed over the chip. Changes in the refractive index at the gold surface as the analyte binds to and dissociates from the ligand, are monitored to generate sensograms from which kon, koff, and Kd can be calculated.
At LifeSensors, we use SPR to study ligand-protein binding for drug discovery by validating hit compounds as a secondary screen. LifeSensors has established a robust protocol to study PROTACs interaction with the target and E3 ligase as both binary and ternary complex formation. With over 30 ligases at our disposal, we have optimized conditions to study interaction with novel binders for PROTAC applications.
LifeSensors SPR studies are performed by highly trained technicians with thorough expertise in SPR and broad biophysics knowledge to guide clients in drug discovery of challenging targets. We use the Bruker Sierra SPR-32 Pro high-throughput SPR analyzer that can enable 13,200 interactions studies (4400 samples) per day. Their breakthrough SPR+ detector allows for superior sensitivity with signal-to-noise ratio of 0.02 RU derived as a function of imaging SPR (SPRi) and high-speed optical scanning. It is crucial to have the best signal-to-noise ratio when screening, identifying, and characterizing novel small molecule ligands. This is key for PROTAC studies to evaluate binary interactions.
Service Highlights
- Tailored solutions to study protein-protein, protein-small molecules/PROTAC interactions
- Characterization of small molecule and PROTACs binding to E3 and target proteins
- Characterization of E3 and DUB ligands and demonstrate selectivity
- High-throughput screening with kinetics and affinity reporting (kon, koff, and Kd)
- Resources
Learn More About Our E3 ELISA Assay Here
Slides to learn about our E3 technologies
Learn More About UbiTest Substrate Validation Here
Quantification ubiquitination
Understanding E3 Ligase Classifications
Important introduction to E3 Ligases and new classifications beyond the standard familiy format (2025)
- Related Products
- E3 Ubiquitin Ligase Assays
