Autophagy is a process by which eukaryotic cells degrade and recycle proteins and organelles using intracellular lysosomes. Malfunctions in autophagy have been shown to induce many diseases including cancer, neurodegeneration, diabetes, autoimmunity, and cardiovascular diseases.
Autophagy can be induced by many intra- and extra-cellular stress conditions such as endoplasmic reticulum stress, oxidative stress, starvation, growth factor depletion, and osmotic stress. It starts with the formation of double membrane autophagosomes, which engulf all the cargos of cytoplasm. The autophagosomes fuse with lysosomes and release the cargos to the highly acidic and hydrolytic environment, which leads to the degradation and recycling of the cellular building blocks across the lysosomal membrane. There are two major types of autophagy: (1) non-selective autophagy (bulk autophagy); and (2) selective autophagy. Selective autophagy is regulated by ubiquitylation on the aggregated cargo proteins.
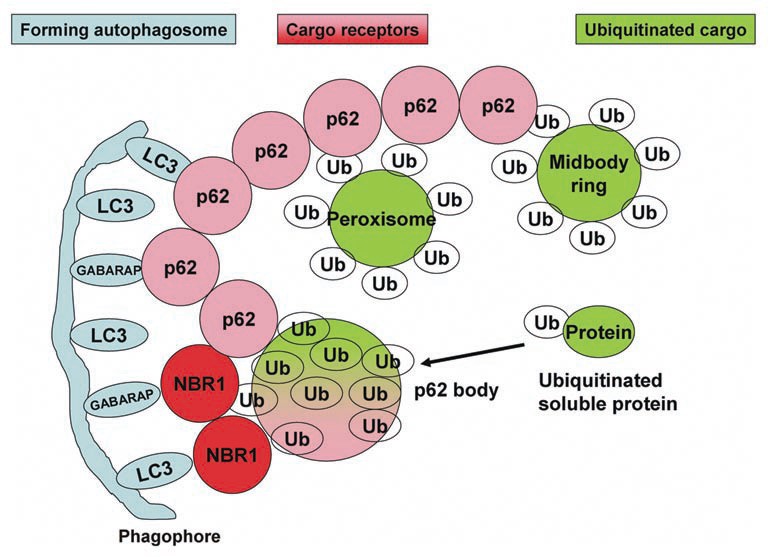
Ubiquitylation plays a critical role in selective autophagy. Attachment of the ubiquitin molecule to normally long-lived cytoplasmic proteins or to organelles has been shown to result in p62-dependent autophagic degradation. p62 and Nbr1, both of which contain an ubiquitin-binding domain at their extreme C-termini, work as cargo adaptors that allow the recognition of K48- or K63-linked polyubiquitylated proteins.
Understanding an E3 ligase is an essential step for choosing a target for screening in the case of a molecular
With ubiquitin’s core functions in plants and animals being so similar it has long been speculated that insights into the
The functional consequences of protein polyubiquitination are determined by the type of ubiquitin chain assembled on the substrate. Among the
While direct K-RAS inhibitors represent a major therapeutic advance, targeting this oncoprotein remains a significant challenge due to limited druggable
Pan-selective Tandem Ubiquitin-Binding Entities (TUBEs) are engineered, high-affinity reagents composed of multiple ubiquitin-associated (UBA) domains that bind polyubiquitin chains with
LifeSensors is pleased to bring attention to some of our academic colleagues who have used our products in the past
Understanding an E3 ligase is an essential step for choosing a target for screening in the case of a molecular
With ubiquitin’s core functions in plants and animals being so similar it has long been speculated that insights into the
The functional consequences of protein polyubiquitination are determined by the type of ubiquitin chain assembled on the substrate. Among the
While direct K-RAS inhibitors represent a major therapeutic advance, targeting this oncoprotein remains a significant challenge due to limited druggable
Pan-selective Tandem Ubiquitin-Binding Entities (TUBEs) are engineered, high-affinity reagents composed of multiple ubiquitin-associated (UBA) domains that bind polyubiquitin chains with
LifeSensors is pleased to bring attention to some of our academic colleagues who have used our products in the past