LifeSensors’ micoarrays can make your ideas a reality-
Services are not limited to those described above. Studies can be extended to characterization of chain linkage specificity to determine which ubiquitin chain linkages your E3 makes and your DUB prefers. LifeSensors has observed that substrate specificity of E3s can change in the presence of a single-lysine mutation of ubiquitin. If you have an experimental question that could be applied to the array, we would be happy to provide a free consultation and develop a customized service for you.
TECHNOLOGY OVERVIEW
A hit on our arrays could lead to your next published paper, funded grant application, or drug target.
Contact us to learn more about this service.
What are the capabilities of Lifesensors’ Ubiquitin Protein Microarray?
Our Microarrays are multifaceted and broad in nature, making use of approximately 20,000 gene products. This versatility provides researchers with a wide variety of capabilites. These include:
- Identifying substrates for deubiquitylating enzymes (DUB)
- Identifying substrates for E3 ligase
- Determining E3 ligases that ubiquitylate your substrate
- Determining DUBs that deubiquitylate your substrate
Why make use of LifeSensors’ Ubiquitin Protein Microarray services?
In addition to the versatile and sensative design of our microarray platform, LifeSensors’ has the most innovative catalog of ubiquitin research tools available on the market today. These research tools in combination with our unique expertise enables our scientists to provide you with a straightforward approach to furthering your research.
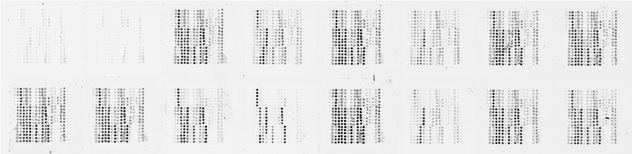
This image of raw microarray data is typical of what is generated from our protein microarray
Human Protein Microarrays
LifeSensors has several commercially available high-density protein microarrays available. Our team stands ready to advise you on which array is most appropriate for your experiment.
A comprehensive list of these microarrays is available here.
References
- Cox E, Uzoma I, Guzzo C, Jeong JS, Matunis M, Blackshaw S, Zhu H. Identification of SUMO E3 ligase-specific substrates using the HuProt human proteome microarray. Methods Mol Biol. 2015;1295:455-63.
- Loch CM, Eddins MJ, Strickler JE. Protein microarrays for the identification of praja1 e3 ubiquitin ligase substrates. Cell Biochem Biophys. 2011 Jun;60(1-2):127-35.
- Loch CM, Strickler JE. A microarray of ubiquitylated proteins for profiling deubiquitylase activity reveals the critical roles of both chain and substrate. Biochim Biophys Acta. 2012 Nov;1823(11):2069-78.
- Merbl Y, Kirschner MW. Post-Translational Modification Profiling–a High-Content Assay for Identifying Protein Modifications in Mammalian Cellular Systems. Curr Protoc Protein Sci. 2014 Aug 1;77:27.8.1-27.8.13.
- Persaud A, Alberts P, Amsen EM, Xiong X, Wasmuth J, Saadon Z, Fladd C, Parkinson J, Rotin D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol Syst Biol. 2009;5:33
