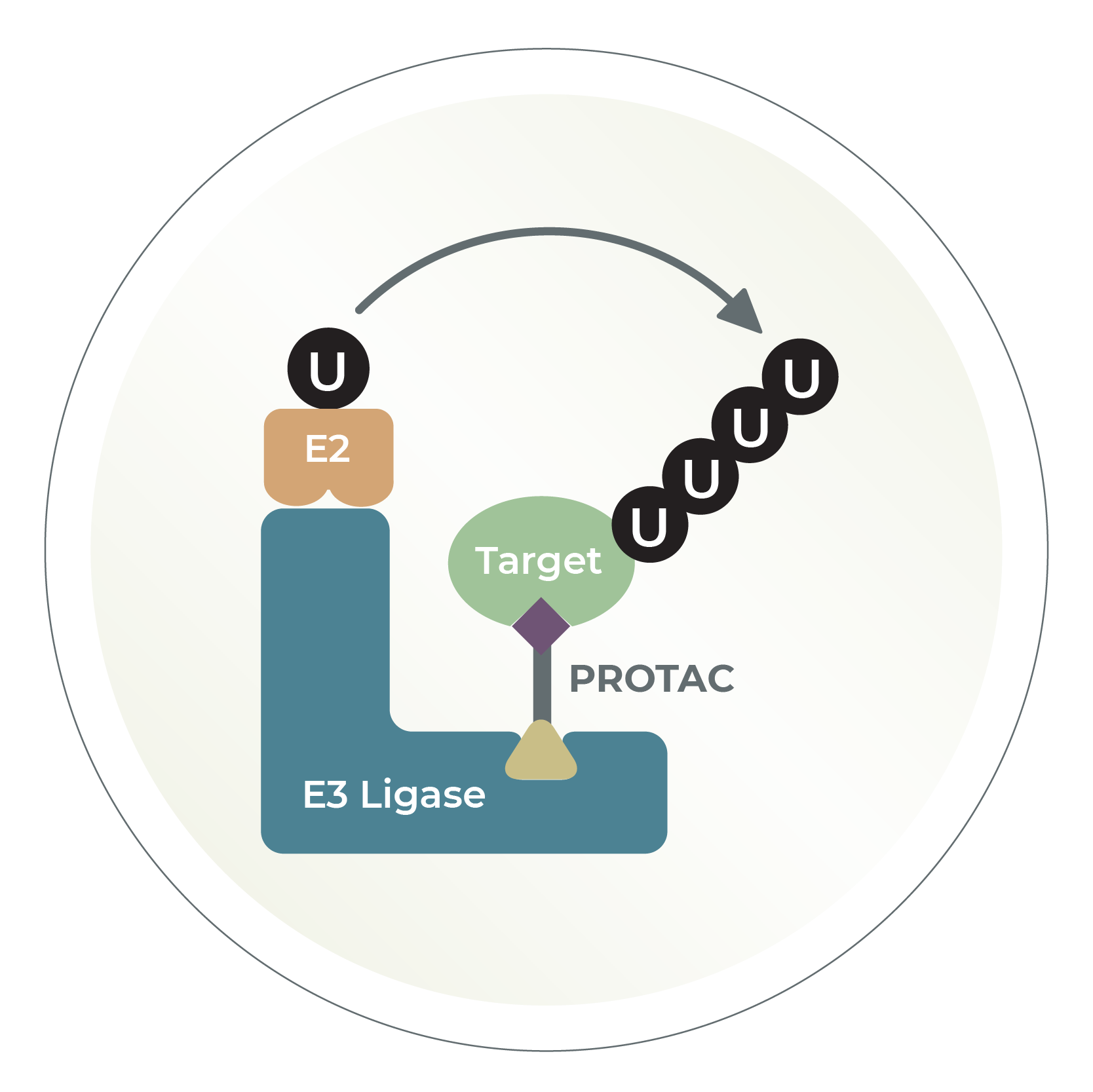
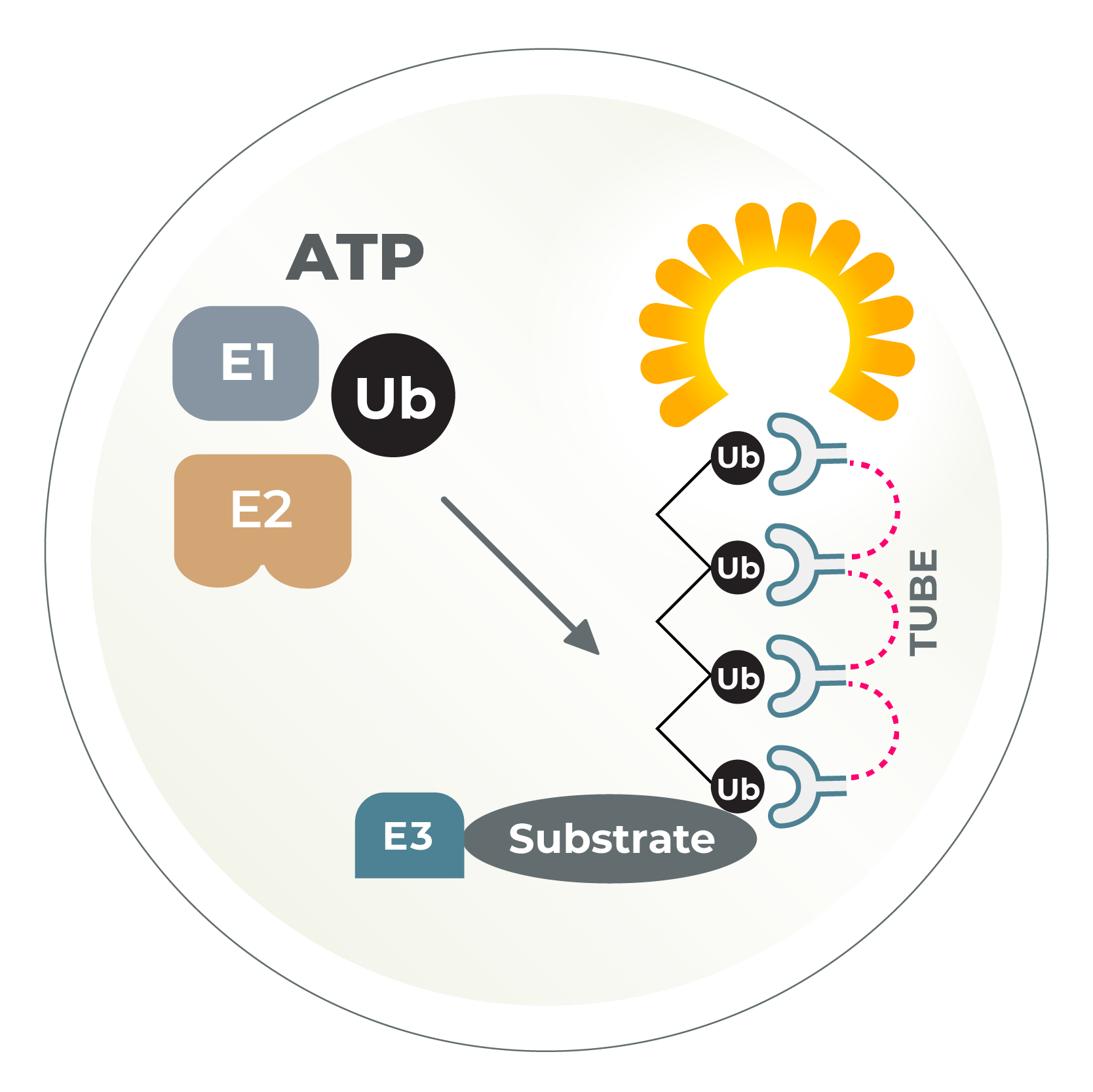
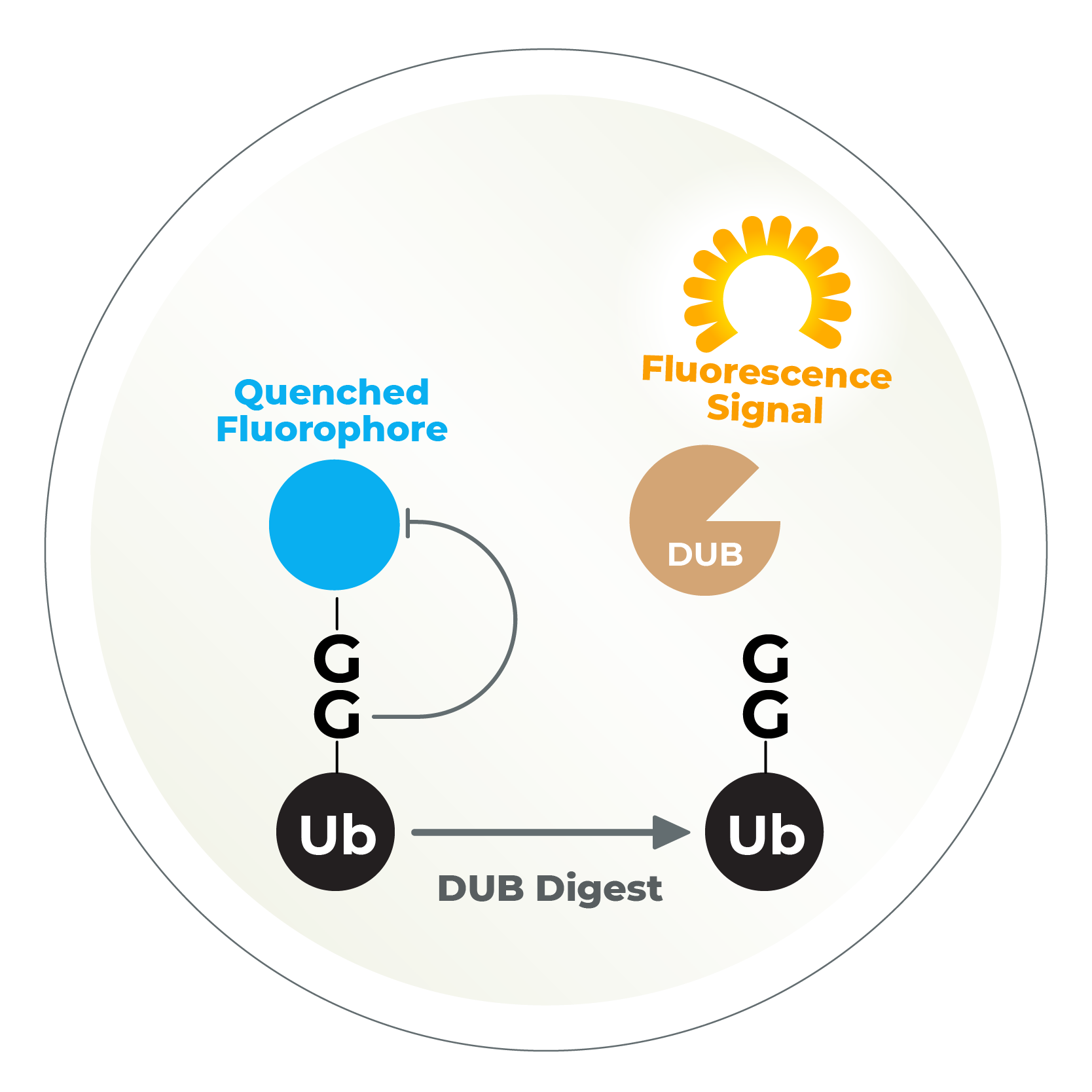
Plate-based assays, best E2 identification, ubiquitin chain identification, compound library screening, custom assays..
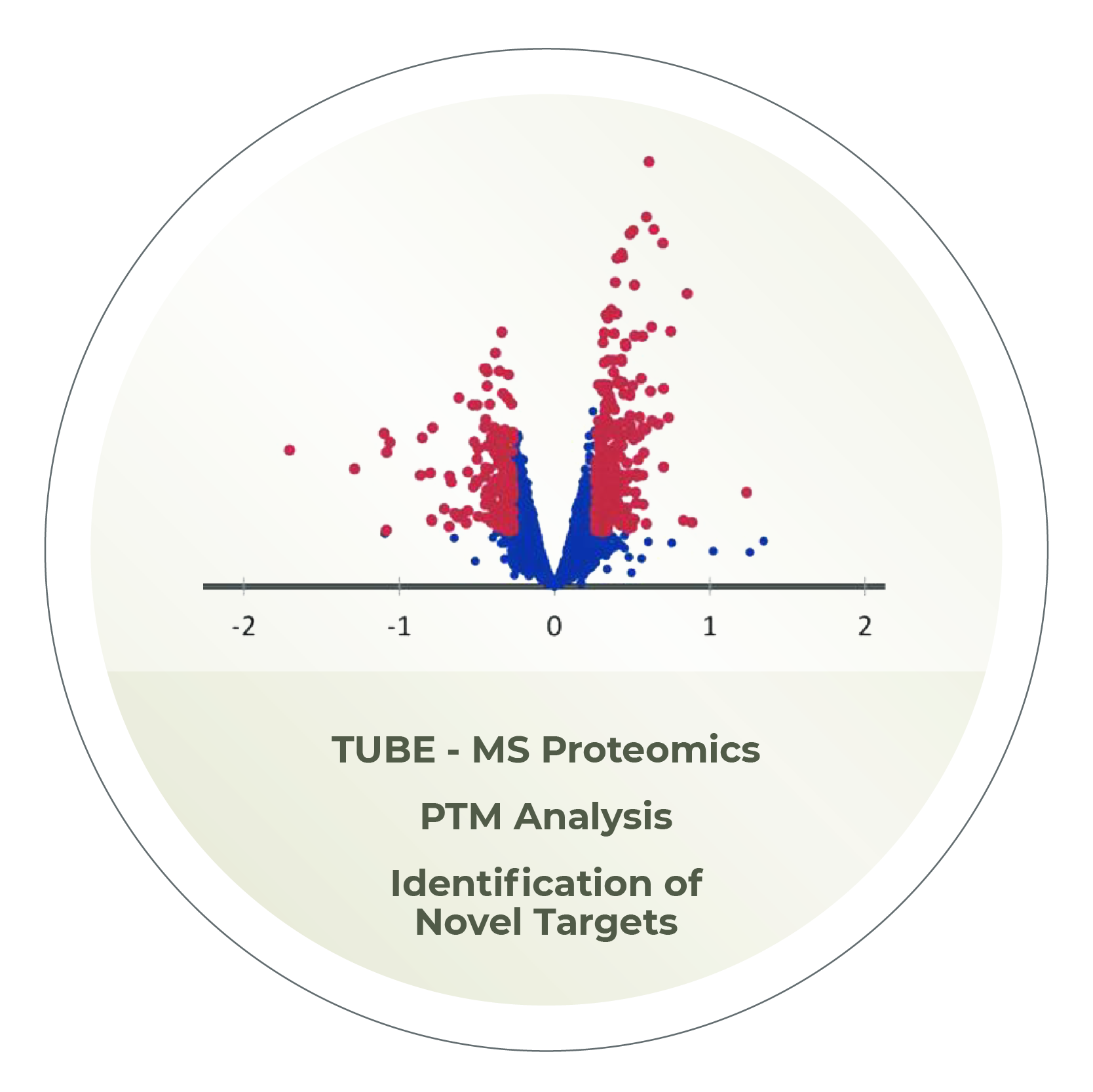
Global substrate ID, TUBE-based affinity matrices, tandem mass spec, and custom assays
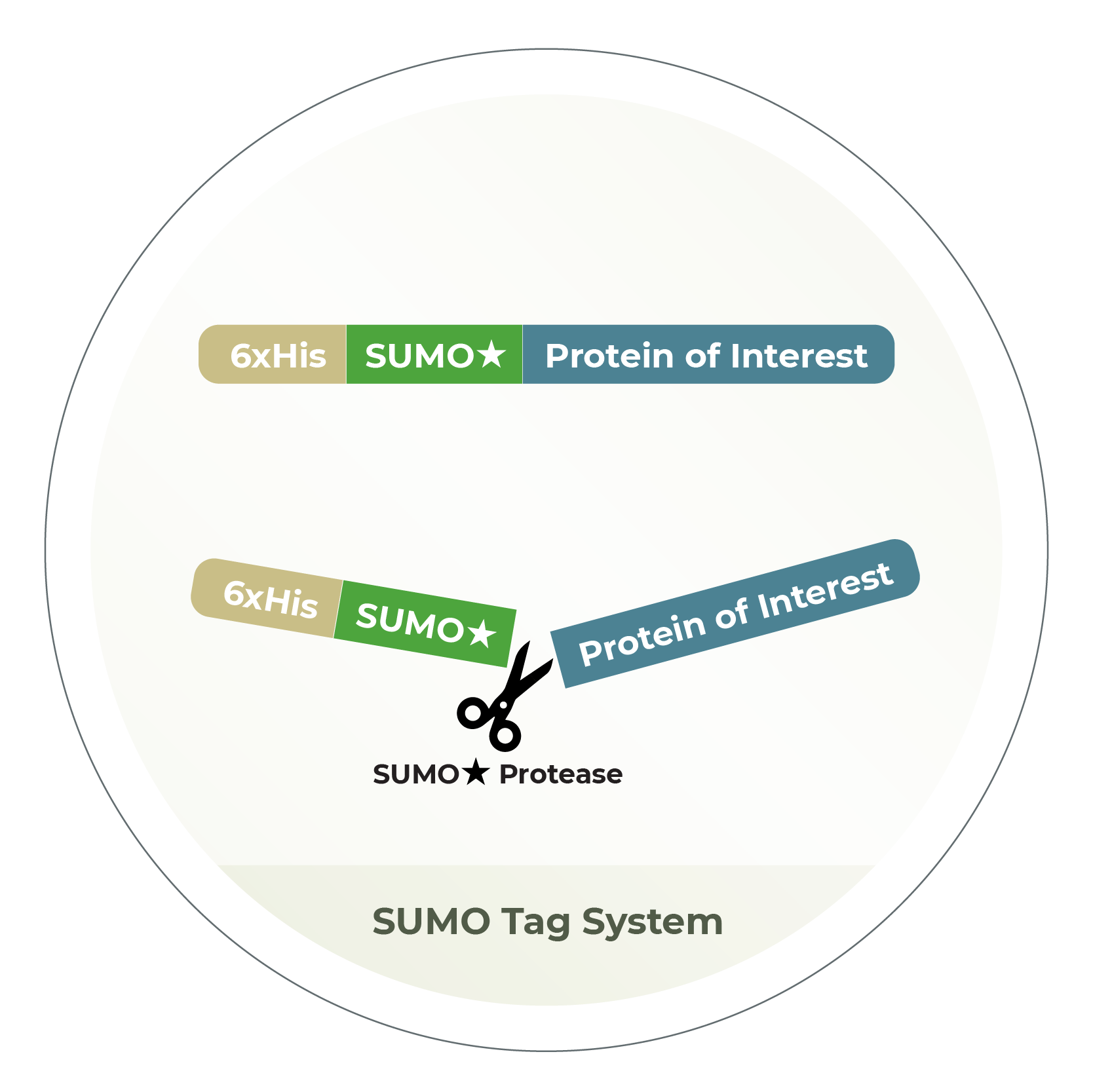
SUMO-tag system for both prokaryotic and eukaryotic cells to maximize protein expression
Plate-based assays, compound library screening, ubiquitin chain selectivity, custom assays